leap_ec package
Subpackages
- leap_ec.binary_rep package
- leap_ec.contrib package
- leap_ec.distrib package
- leap_ec.executable_rep package
- Submodules
- leap_ec.executable_rep.cgp module
- leap_ec.executable_rep.executable module
- leap_ec.executable_rep.neural_network module
- leap_ec.executable_rep.problems module
- leap_ec.executable_rep.rules module
PittRulesDecoderPittRulesDecoder.action_boundsPittRulesDecoder.bounds()PittRulesDecoder.condition_boundsPittRulesDecoder.decode()PittRulesDecoder.genome_to_rules()PittRulesDecoder.initializer()PittRulesDecoder.mutator()PittRulesDecoder.num_genes_per_rulePittRulesDecoder.num_inputsPittRulesDecoder.num_memory_registersPittRulesDecoder.num_outputs
PittRulesExecutablePlotPittRuleProbeRule
- Module contents
- leap_ec.int_rep package
- leap_ec.landscape_features package
- leap_ec.multiobjective package
- Submodules
- leap_ec.multiobjective.asynchronous module
- leap_ec.multiobjective.nsga2 module
- leap_ec.multiobjective.ops module
- leap_ec.multiobjective.probe module
- leap_ec.multiobjective.problems module
- Module contents
- leap_ec.real_rep package
- Submodules
- leap_ec.real_rep.initializers module
- leap_ec.real_rep.ops module
- leap_ec.real_rep.problems module
AckleyProblemCosineFamilyProblemGaussianProblemGriewankProblemLangermannProblemLunacekProblemMatrixTransformedProblemNoisyQuarticProblemParabaloidProblemQuadraticFamilyProblemRastriginProblemRosenbrockProblemScaledProblemSchwefelProblemShekelProblemSpheroidProblemStepProblemTranslatedProblemWeierstrassProblemplot_2d_contour()plot_2d_function()plot_2d_problem()random()random_orthonormal_matrix()
- Module contents
- leap_ec.segmented_rep package
Submodules
leap_ec.algorithm module
Provides convenient monolithic functions that wrap a lot of common function- ality.
generational_ea() for a typical generational model
multi_population_ea() for invoking an EA using sub-populations
random_search() for a more naive strategy
- leap_ec.algorithm.generational_ea(max_generations: int, pop_size: int, problem, representation, pipeline, stop=<function <lambda>>, init_evaluate=<bound method Individual.evaluate_population of <class 'leap_ec.individual.Individual'>>, k_elites: int = 1, start_generation: int = 0, context={'leap': {'distrib': {'non_viable': 0}}})
This function provides an evolutionary algorithm with a generational population model.
When called this initializes and evaluates a population of size pop_size using the init_evaluate function and then pipes it through an operator pipeline (i.e. a list of operators) to obtain offspring. Wash, rinse, repeat.
The algorithm is provided here at the “metaheuristic” level: in order to apply it to a particular problem, you must parameterize it with implementations of its various components. You must decide the population size, how individuals are represented and initialized, the pipeline of reproductive operators, etc. A metaheuristic template of this kind can be used to implement genetic algorithms, genetic programming, certain evolution strategies, and all manner of other (novel) algorithms by passing in appropriate components as parameters.
- Parameters
max_generations (int) – The max number of generations to run the algorithm for. Can pass in float(‘Inf’) to run forever or until the stop condition is reached.
pop_size (int) – Size of the initial population
stop (int) – A function that accepts a population and returns True iff it’s time to stop evolving.
problem (Problem) – the Problem that should be used to evaluate individuals’ fitness
representation – How the problem is represented in individuals
pipeline (list) – a list of operators that are applied (in order) to create the offspring population at each generation
init_evaluate – a function used to evaluate the initial population, before the main pipeline is run. The default of Individual.evaluate_population is suitable for many cases, but you may wish to pass a different operator in for distributed evaluation or other purposes.
k_elites – keep k elites
start_generation – index of the first generation to count from (defaults to 0). You might want to change this, for example, in experiments that involve stopping and restarting an algorithm.
- Returns
the final population
The intent behind this kind of EA interface is to allow the complete configuration of a basic evolutionary algorithm to be defined in a clean and readable way. If you define most of the components in-line when passing them to the named arguments, then the complete configuration of an algorithmic experiment forms one concise code block. Here’s what a basic (mu, lambda)-style EA looks like (that is, an EA that throws away the parents at each generation in favor of their offspring):
>>> from leap_ec import Individual, Representation >>> from leap_ec.algorithm import generational_ea, stop_at_generation >>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.binary_rep.initializers import create_binary_sequence >>> from leap_ec.binary_rep.ops import mutate_bitflip >>> import leap_ec.ops as ops >>> pop_size = 5 >>> final_pop = generational_ea(max_generations=100, pop_size=pop_size, ... ... problem=MaxOnes(), # Solve a MaxOnes Boolean optimization problem ... ... representation=Representation( ... initialize=create_binary_sequence(length=10) # Initial genomes are random binary sequences ... ), ... ... # The operator pipeline ... pipeline=[ ... ops.tournament_selection, # Select parents via tournament selection ... ops.clone, # Copy them (just to be safe) ... mutate_bitflip(expected_num_mutations=1), # Basic mutation with a 1/L mutation rate ... ops.UniformCrossover(p_swap=0.4), # Crossover with a 40% chance of swapping each gene ... ops.evaluate, # Evaluate fitness ... ops.pool(size=pop_size) # Collect offspring into a new population ... ])
The algorithm runs immediately and returns the final population:
>>> print(*final_pop, sep='\n') Individual<...> ... Individual<...> ... Individual<...> ... ... Individual<...> ...
You can get the best individual by using max (since comparison on individuals is based on the Problem associated with them, this will return the best individaul even on minimization problems):
>>> max(final_pop) Individual<...>...
- leap_ec.algorithm.multi_population_ea(max_generations, num_populations, pop_size, problem, representation, shared_pipeline, subpop_pipelines=None, stop=<function <lambda>>, init_evaluate=<bound method Individual.evaluate_population of <class 'leap_ec.individual.Individual'>>, context={'leap': {'distrib': {'non_viable': 0}}})
An EA that maintains multiple (interacting) subpopulations, i.e. for implementing island models.
This effectively executes several EAs concurrently that share the same generation counter, and which share the same representation (
Individual,Decoder) and objective function (Problem), and which share all or part of the same operator pipeline.- Parameters
max_generations (int) – The max number of generations to run the algorithm for. Can pass in float(‘Inf’) to run forever or until the stop condition is reached.
num_populations (int) – The number of separate populations to maintain.
pop_size (int) – Size of each initial subpopulation
stop (int) – A function that accepts a list of populations and returns True iff it’s time to stop evolving.
problem (Problem) – the Problem that should be used to evaluate individuals’ fitness
representation – the Representation that governs the creation and decoding of individuals. If a list of Representation objects is given, then different representations will be used for different subpopulations; else the same representation will be used for all subpopulations.
shared_pipeline (list) – a list of operators that every population will use to create the offspring population at each generation
subpop_pipelines (list) – a list of population-specific operator lists, the ith of which will only be applied to the ith population (after the shared_pipeline). Ignored if None.
init_evaluate – a function used to evaluate the initial population, before the main pipeline is run. The default of Individual.evaluate_population is suitable for many cases, but you may wish to pass a different operator in for distributed evaluation or other purposes.
- Returns
a list of lists of each of the subpopulations.
To turn a multi-population EA into an island model, use the
leap_ec.ops.migrate()operator in the shared pipeline. This operator takes a NetworkX graph describing the topology of connections between islands as input.For example, here’s how we might define a fully connected 4-island model that solves a
leap_ec.real_rep.problems.SchwefelProblemusing a real-vector representation:>>> import networkx as nx >>> from leap_ec.algorithm import multi_population_ea >>> from leap_ec import ops >>> from leap_ec.real_rep.ops import mutate_gaussian >>> from leap_ec.real_rep import problems >>> from leap_ec.decoder import IdentityDecoder >>> from leap_ec.representation import Representation >>> from leap_ec.real_rep.initializers import create_real_vector >>> >>> topology = nx.complete_graph(4) >>> nx.draw_networkx(topology, with_labels=True) >>> problem = problems.SchwefelProblem(maximize=False) ... >>> l = 2 # Length of the genome >>> pop_size = 10 >>> pops = multi_population_ea(max_generations=10, ... num_populations=topology.number_of_nodes(), ... pop_size=pop_size, ... ... problem=problem, ... ... representation=Representation( ... individual_cls=Individual, ... decoder=IdentityDecoder(), ... initialize=create_real_vector(bounds=[problem.bounds] * l) ... ), ... ... shared_pipeline=[ ... ops.tournament_selection, ... ops.clone, ... mutate_gaussian(std=30, ... expected_num_mutations='isotropic', ... bounds=problem.bounds), ... ops.evaluate, ... ops.pool(size=pop_size), ... ops.migrate(topology=topology, ... emigrant_selector=ops.tournament_selection, ... replacement_selector=ops.random_selection, ... migration_gap=5) ... ]) >>> pops [[Individual<...>(...), ..., Individual<...>(...)], ..., [Individual<...>(...), ..., Individual<...>(...)]]
We can now run the algorithm by pulling output from its generator, which gives us the best individual in each population at each generation:
While each population is executing, multi_population_ea writes the index of the current subpopulation to context[‘leap’][ ‘subpopulation’]. That way shared operators (such as
leap.ops.migrate()) have the option of accessing the share context to learn which subpopulation they are currently working with.TODO find a way to use Dask to parallelize populations, likely by having a Dask worker for each sub-poplulation.
- leap_ec.algorithm.random_search(evaluations, problem, representation, pipeline, context={'leap': {'distrib': {'non_viable': 0}}})
This function performs random search of a solution space using the given representation and problem.
Random search is often used as a control in evolutionary algorithm experiments: if your pet algorithm can’t perform better than random search, then it’s a sign that you’ve barked up the wrong tree!
This implementation also allows you to pass in an operator pipeline, which will be applied to each individual. The pipeline must have the following types of operators:
- a selection operator, probably cyclic_selection since there will be only
one individual from which to choose
clone operator to ensure we don’t overwrite the previous individual
a pertubation operator, likely a simple mutation pipeline operator
evaluate operator so we know where the new individual is in the space
pool(size=1) to act as a pipeline sink to pull the new individuals through
- Parameters
evaluations – how many evaluations to perform
problem – the Problem instance to use for evaluating individuals
representation – the Representation describing individuals
pipeline – reproductive operator pipeline
context – optional context for storing state as algorithm progresses
- Returns
the series of individuals that describe a random walk
>>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.binary_rep.initializers import create_binary_sequence >>> from leap_ec.binary_rep.ops import mutate_bitflip >>> from leap_ec.decoder import IdentityDecoder >>> from leap_ec.representation import Representation >>> from leap_ec.individual import Individual >>> from leap_ec.ops import evaluate, clone, cyclic_selection, pool >>> result = random_search(evaluations=5, ... problem=MaxOnes(), # Solve a MaxOnes Boolean optimization problem ... ... representation=Representation( ... individual_cls=Individual, # Use the standard Individual as the prototype for the population ... decoder=IdentityDecoder(), # Genotype and phenotype are the same for this task ... initialize=create_binary_sequence(length=3) # Initial genomes are random binary sequences ... ), ... pipeline=[cyclic_selection, ... clone, ... mutate_bitflip(expected_num_mutations=3), ... evaluate, ... pool(size=1)]) >>> assert(len(result) == 5)
The algorithm outputs a list containing all the generated individuals.
- leap_ec.algorithm.stop_at_generation(max_generation: int, context={'leap': {'distrib': {'non_viable': 0}}})
A stopping criterion function that checks the ‘generation’ count in the context object and returns True iff it is >= max_generation.
The resulting function takes a population argument, which is ignored.
For example:
>>> from leap_ec import context >>> stop = stop_at_generation(100)
If we set the generation field in the context object (this value will typically be updated by the algorithm as it runs) like so:
>>> context['leap']['generation'] = 15
Then we don’t stop yet:
>>> stop(population=[]) False
We do stop at the 100th generation:
>>> context['leap']['generation'] = 100 >>> stop([]) True
leap_ec.data module
A module for synthetic data that we use in test and examples.
leap_ec.decoder module
Defines the Decoder base class.
Decoders are used to translate from genotypic to phenotypic space. E.g., binary strings may have to be decoded into corresponding integers or real values meaningful to a Problem.
- class leap_ec.decoder.Decoder
Bases:
ABC- Decoders in LEAP implement how solutions to a problem are represented.
Specifically, a
Decoderconverts anIndividual’s genotype (which is a format that can easily be manipulated by mutation and recombination operators) into a phenotype (which is a format that can be fed directly into aProblemobject to obtain a fitness value).
Genotypes and phenotypes can be of arbitrary type, from a simple list of numbers to a complex data structure. Choosing a good genotypic representation and genotype-to-phenotype mapping for a given problem domain is a critical part of evolutionary algorithm design: the
Decoderobject that an algorithm uses can have a big impact on the effectiveness of your metaheuristics.In LEAP, a
Decoderis typically used byIndividualas an intermediate step in calculating its own fitness.For example, say that we want to use a binary-represented
Individualto solve a real-valued optimization problem, such asSchwefelProblem. Here, the genotype is a vector of binary values, whereas the phenotype is its corresponding float vector.We can use a
BinaryToIntDecoderto express this mapping. And when we initialize an individual, we give it all three pieces of this information:>>> from leap_ec.binary_rep.decoders import BinaryToRealDecoder >>> from leap_ec.individual import Individual >>> from leap_ec.real_rep.problems import SchwefelProblem >>> import numpy as np >>> genome = np.array([0, 1, 1, 0, 1, 0, 1, 1]) >>> decoder = BinaryToRealDecoder((4, -5.12, 5.12), (4, -5.12, 5.12)) # Every 4 bits map to a float on (-5.12, 5.12) >>> ind = Individual(genome, decoder=decoder, problem=SchwefelProblem())
Now we can decode the individual to examine its phenotype:
>>> ind.decode() array([-1.024 , 2.38933333])
This call is just a wrapper for the
Decoder, which has the same output:>>> decoder.decode(genome) array([-1.024 , 2.38933333])
But now
Individualalso has everything it needs to evaluate its own fitness:>>> ind.evaluate() 836.4453949...
Calling evaluate() also has the side effect of setting the fitness attribute:
>>> ind.fitness 836.4453949...
- abstract decode(genome, *args, **kwargs)
- Parameters
genome – a genome you wish to convert
- Returns
the phenotype associated with that genome
- class leap_ec.decoder.IdentityDecoder
Bases:
DecoderA decoder that maps a genome to itself. This acts as a ‘direct’ or ‘phenotypic’ encoding: Use this when your genotype and phenotype are the same thing.
- decode(genome, *args, **kwargs)
- Returns
the input genome.
For example:
>>> import numpy as np >>> d = IdentityDecoder() >>> d.decode(np.array([0.5, 0.6, 0.7])) array([0.5, 0.6, 0.7])
leap_ec.distrib module
leap_ec.global_vars module
This defines a global context that is a dictionary of dictionaries. The intent is for certain operators and functions to add to and modify this context. Third party operators and functions will just add a new top-level dedicated key.
context[‘leap’] is for storing general LEAP running state, such as current generation.
context[‘leap’][‘distrib’] is for storing leap.distrib running state
context[‘leap’][‘distrib’][‘non_viable’] accumulates counts of non-viable individuals during distrib.eval_pool() and distrib.async_eval_pool() runs.
leap_ec.individual module
Defines Individual
- class leap_ec.individual.Individual(genome, decoder=IdentityDecoder(), problem=None)
Bases:
objectRepresents a single solution to a Problem.
We represent an Individual by a genome and a fitness. Individual also maintains a reference to the Problem it will be evaluated on, and an decoder, which defines how genomes are converted into phenomes for fitness evaluation.
- clone()
Create a ‘clone’ of this Individual, copying the genome, but not fitness.
The fitness of the clone is set to None. A new UUID is generated and assigned to sefl.uuid. The parents set is updated to include the UUID of the parent. A shallow copy of the parent is made, too, so that ancillary state is also copied.
A deep copy of the genome will be created, so if your Individual has a custom genome type, it’s important that it implements the __deepcopy__() method.
>>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.decoder import IdentityDecoder >>> import numpy as np >>> genome = np.array([0, 1, 1, 0]) >>> ind = Individual(genome, IdentityDecoder(), MaxOnes()) >>> ind_copy = ind.clone() >>> ind_copy.genome == ind.genome array([ True, True, True, True]) >>> ind_copy.problem == ind.problem True >>> ind_copy.decoder == ind.decoder True
- classmethod create_population(n, initialize, decoder, problem)
A convenience method for initializing a population of the appropriate subtype.
- Parameters
n – The size of the population to generate
initialize – A function f(m) that initializes a genome
decoder – The decoder to attach individuals to
problem – The problem to attach individuals to
- Returns
A list of n individuals of this class’s (or subclass’s) type
- decode(*args, **kwargs)
Determine the indivdual’s phenome.
This is done by passing the genome self.decoder.
The result is both returned and saved to self.phenome.
- Returns
the decoded value for this individual
- evaluate()
determine this individual’s fitness
This is done by outsourcing the fitness evaluation to the associated Problem object since it “knows” what is good or bad for a given phenome.
- See also
ScalarProblem.worse_than
- Returns
the calculated fitness
- evaluate_imp()
This is the evaluate ‘implementation’ called by self.evaluate(). It’s intended to be optionally over-ridden by sub-classes to give an opportunity to pass in ancillary data to the evaluate process either by tailoring the problem interface or that of the given decoder.
- classmethod evaluate_population(population)
Convenience function for bulk serial evaluation of a given population
- Parameters
population – to be evaluated
- Returns
evaluated population
- property phenome
If the phenome has not yet been decoded, do so.
- class leap_ec.individual.RobustIndividual(genome, decoder=IdentityDecoder(), problem=None)
Bases:
IndividualThis adds exception handling for evaluations
After evaluation self.is_viable is set to True if all went well. However, if an exception is thrown during evaluation, the following happens:
self.is_viable is set to False
self.fitness is set to math.nan
self.exception is assigned the exception
- evaluate()
determine this individual’s fitness
Note that if an exception is thrown during evaluation, the fitness is set to NaN and self.is_viable to False; also, the returned exception is assigned to self.exception for possible later inspection. If the individual was successfully evaluated, self.is_viable is set to true. NaN fitness values will figure into comparing individuals in that NaN will always be considered worse than non-NaN fitness values.
- Returns
the calculated fitness
- class leap_ec.individual.WholeEvaluatedIndividual(genome, decoder=IdentityDecoder(), problem=None)
Bases:
IndividualAn Individual that, when evaluated, passes its whole self to the evaluation function, rather than just its phenome.
In most applications, fitness evaluation requires only phenome information, so that is all that we pass from the Individual to the Problem. This is important, because during distributed evaluation, we want to pass as little information as possible across nodes.
WholeEvaluatedIndividual is used for special cases where fitness evaluation needs access to more information about an individual than its phenome. This is strange in most cases and should be avoided, but can make certain algorithms more elegant (ex. it’s helpful when interpreting cooperative coevolution as an island model).
This can dramatically slow down distributed evaluation (i.e. with dask) in some applications because the entire individual will be sent over a TCP/IP connection instead of just the phenome, so use with caution.
- evaluate_imp()
This is the evaluate ‘implementation’ called by self.evaluate(). It’s intended to be optionally over-ridden by sub-classes to give an opportunity to pass in ancillary data to the evaluate process either by tailoring the problem interface or that of the given decoder.
leap_ec.multiobjective module
leap_ec.ops module
Fundamental evolutionary operators.
This module provides many of the most important functions that we string together to create EAs out of operator pipelines. You’ll find many traditional selection and reproduction strategies here, as well as components for classic algorithms like island models and cooperative coevolution.
Representation-specific operators tend to reside within their own subpackages,
rather than here. See for example leap_ec.real_rep.ops and
leap_ec.binary_rep.ops.
- class leap_ec.ops.CooperativeEvaluate(num_trials: int, collaborator_selector, log_stream=None, combine=<function concat_combine>, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
OperatorA simple, non-parallel implementation of cooperative coevolutionary fitness evaluation.
- Parameters
num_trials (int) – the number of combined solutions & fitness estimates to collect when computing a partial solution’s fitness.
collaborator_selector – a selection operator that we use to choose individuals from the other subpopulations to create a combined solution.
context – the algorithm’s state context. Used to access subpopulation information.
log_stream – optional file object to collect statistics about combined individuals to.
combine – the function used to combine partial solutions into combined solutions.
- class leap_ec.ops.Crossover(persist_children, p_xover)
Bases:
Operator- abstract recombine(parent_a, parent_b)
Perform recombination between two parents to produce two new individuals.
- class leap_ec.ops.NAryCrossover(num_points=2, p_xover=1.0, persist_children=False)
Bases:
CrossoverDo crossover between individuals between N crossover points.
1 < n < genome length - 1
We also assume that the passed in individuals are clones of parents.
>>> from leap_ec.individual import Individual >>> from leap_ec.ops import NAryCrossover >>> import numpy as np
>>> genome1 = np.array([0, 0]) >>> genome2 = np.array([1, 1]) >>> first = Individual(genome1) >>> second = Individual(genome2) >>> pop = [first, second] >>> select = naive_cyclic_selection(pop)
>>> op = NAryCrossover() >>> result = op(select)
>>> new_first = next(result) >>> new_second = next(result)
If persist_children is True and there is a child that was made by crossover but isn’t used in the first call, it will be yielded in a future call.
>>> op = NAryCrossover(p_xover=0.0, persist_children=True) >>> >>> next(op(select)) is first # Create an iterator loop with op(select) and consume 1 individual True >>> next(op(select)) is second # Create a different iterator loop with op(select) True
With persist_children set to False, the second child will not be yielded if the iterator is consumed an odd number of times. Instead, on the next call the loop is started anew.
>>> op = NAryCrossover(p_xover=0.0, persist_children=False) >>> >>> next(op(select)) is first # Create an iterator loop with op(select) and consume 1 individual True >>> next(op(select)) is second # Create a different iterator loop with op(select) False
- Parameters
num_points – how many crossing points do we use? Defaults to 2, since 2-point crossover has been shown to be the least disruptive choice for this value.
p – the probability that crossover is performed.
persist_children (bool) – whether unyielded children should persist between calls. This is useful for leap_ec.distrib.asynchronous.steady_state, where the pipeline may only produce one individual at a time.
- Returns
a pipeline operator that returns two recombined individuals (with probability p), or two unmodified individuals (with probability 1 - p)
- recombine(parent_a, parent_b)
Perform recombination between two parents to produce two new individuals.
- class leap_ec.ops.Operator
Bases:
ABCAbstract base class that documents the interface for operators in a LEAP pipeline.
LEAP treats operators as functions of two arguments: the population, and a “context” dict that may be used in some algorithms to maintain some global state or parameters independent of the population.
TODO The above description is outdated. –Siggy TODO Also this is for a population based operator. We also have operators for individuals
You can inherit from this class to define operators as classes. Classes support operators that take extra arguments at construction time (such as a mutation rate) and maintain some internal private state, and they allow certain special patterns (such as multi-function operators).
But inheriting from this class is optional. LEAP can treat any callable object that takes two parameters as an operator. You may define your custom operators as closures (which also allow for construction-time arguments and internal state), as simple functions ( when no additional arguments are necessary), or as curried functions ( i.e. with the help of toolz.curry(…).
- class leap_ec.ops.UniformCrossover(p_swap: float = 0.2, p_xover: float = 1.0, persist_children=False)
Bases:
CrossoverParameterized uniform crossover iterates through two parents’ genomes and swaps each of their genes with the given probability.
In a classic paper, De Jong and Spears showed that this operator works particularly well when the swap probability p_swap is set to about 0.2. LEAP thus uses this value as its default.
De Jong, Kenneth A., and W. Spears. “On the virtues of parameterized uniform crossover.” Proceedings of the 4th international conference on genetic algorithms. Morgan Kaufmann Publishers, 1991.
>>> from leap_ec.individual import Individual >>> from leap_ec.ops import UniformCrossover, naive_cyclic_selection >>> import numpy as np
>>> genome1 = np.array([0, 0]) >>> genome2 = np.array([1, 1]) >>> first = Individual(genome1) >>> second = Individual(genome2) >>> pop = [first, second] >>> select = naive_cyclic_selection(pop) >>> op = UniformCrossover() >>> result = op(select) >>> new_first = next(result) >>> new_second = next(result)
The probability can be tuned via the p_swap parameter: >>> op = UniformCrossover(p_swap=0.1) >>> result = op(select)
If persist_children is True and there is a child that was made by crossover but isn’t used in the first call, it will be yielded in a future call.
>>> op = UniformCrossover(p_xover=0.0, persist_children=True) >>> >>> next(op(select)) is first # Create an iterator loop with op(select) and consume 1 individual True >>> next(op(select)) is second # Create a different iterator loop with op(select) True
With persist_children set to False, the second child will not be yielded if the iterator is consumed an odd number of times. Instead, on the next call the loop is started anew.
>>> op = UniformCrossover(p_xover=0.0, persist_children=False) >>> >>> next(op(select)) is first # Create an iterator loop with op(select) and consume 1 individual True >>> next(op(select)) is second # Create a different iterator loop with op(select) False
- Parameters
p_swap – how likely are we to swap each pair of genes when crossover is performed
p_xover (float) – the probability that crossover is performed in the first place
persist_children (bool) – whether unyielded children should persist between calls. This is useful for leap_ec.distrib.asynchronous.steady_state, where the pipeline may only produce one individual at a time.
- Returns
a pipeline operator that returns two recombined individuals (with probability p_xover), or two unmodified individuals (with probability 1 - p_xover)
- recombine(parent_a, parent_b)
Perform recombination between two parents to produce two new individuals.
- leap_ec.ops.clone(next_individual: Iterator = '__no__default__') Iterator
clones and returns the next individual in the pipeline
The clone’s fitness is set to None, its parents are set to the individual from which it was cloned (i.e., the parent), and it is assigned its own UUID.
>>> from leap_ec.individual import Individual >>> import numpy as np
Create a common decoder and problem for individuals.
>>> genome = np.array([1, 1]) >>> original = Individual(genome)
>>> cloned_generator = clone(iter([original]))
- Parameters
next_individual – iterator for next individual to be cloned
- Returns
copy of next_individual
- leap_ec.ops.compute_expected_probability(expected_num_mutations: float, individual_genome: List) float
Computed the probability of mutation based on the desired average expected mutation and genome length.
The equation here is \(p = 1/L * \texttt{expected_num_mutations}\). To see why this is correct, note that the number of mutations performed is characterized by a binomial distribution with \(n=L\) trials (one weighted “coin flip” per gene), and that the mean (expected_num_mutations) of a binomial distribution is given by \(n*p = expected_num_mutations\).
- Parameters
expected_num_mutations – times individual is to be mutated on average
individual_genome – genome for which to compute the probability
- Returns
the corresponding probability of mutation
- leap_ec.ops.compute_population_values(population: ~typing.List, offset=0, exponent: int = 1, key=<function <lambda>>) ndarray
Returns a list of values where the zero-point of the population is shifted and the values are scaled by exponentiation.
- Parameters
population – the population to compute values from.
offset – the offset from zero. Specifying offset=’pop-min’ will use the population’s minimum value as the new zero-point. Defaults to 0.
exponent (int) – the power to which values are raised to. Defaults to 1.
key – a function that computes a metric based on an Individual.
- Returns
a numpy array of values that have been shifted by offset and scaled by exponent corresponding to each individual in the population.
- leap_ec.ops.concat_combine(collaborators)
Combine a list of individuals by concatenating their genomes.
You can choose whether this or some other function is used for combining collaborators by passing it into the CooperativeEvaluate constructor.
- leap_ec.ops.const_evaluate(population: List = '__no__default__', value='__no__default__') List
An evaluator that assigns a constant fitness to every individual.
This ignores the Problem associated with each individual for the purpose of assigning a constant fitness.
This is useful for algorithms that need to assign an arbitrary initial fitness value before using their normal evaluation method. Some forms of cooperative coevolution are an example.
- leap_ec.ops.cyclic_selection(population: List = '__no__default__') Iterator
Deterministically returns individuals in order, then shuffles the test_sequence, returns the individuals in that new order, and repeats this process.
>>> from leap_ec.individual import Individual >>> from leap_ec.ops import cyclic_selection >>> import numpy as np
>>> pop = [Individual(np.array([0, 0])), ... Individual(np.array([0, 1]))]
>>> cyclic_selector = cyclic_selection(pop)
- Parameters
population – from which to select
- Returns
the next selected individual
- leap_ec.ops.elitist_survival(offspring: List = '__no__default__', parents: List = '__no__default__', k: int = 1, key=None) List
This allows k best parents to compete with the offspring.
>>> from leap_ec.individual import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> import numpy as np
First, let’s make a “pretend” population of parents using the MaxOnes problem.
>>> pretend_parents = [Individual(np.array([0, 0, 0]), problem=MaxOnes()), ... Individual(np.array([1, 1, 1]), problem=MaxOnes())]
Then a “pretend” population of offspring. (Pretend in that we’re pretending that the offspring came from the parents.)
>>> pretend_offspring = [Individual(np.array([0, 0, 0]), problem=MaxOnes()), ... Individual(np.array([1, 1, 0]), problem=MaxOnes()), ... Individual(np.array([1, 0, 1]), problem=MaxOnes()), ... Individual(np.array([0, 1, 1]), problem=MaxOnes()), ... Individual(np.array([0, 0, 1]), problem=MaxOnes())]
We need to evaluate them to get their fitness to sort them for elitist_survival.
>>> pretend_parents = Individual.evaluate_population(pretend_parents) >>> pretend_offspring = Individual.evaluate_population(pretend_offspring)
This will take the best parent, which has [1,1,1], and replace the worst offspring, which has [0,0,0] (because this is the MaxOnes problem) >>> survivors = elitist_survival(pretend_offspring, pretend_parents)
>>> assert pretend_parents[1] in survivors # yep, best parent is there >>> assert pretend_offspring[0] not in survivors # worst guy isn't
We orginally ordered 5 offspring, so that’s what we better have. >>> assert len(survivors) == 5
Please note that the literature has a number of variations of elitism and other forms of overlapping generations. For example, this may be a good starting point:
De Jong, Kenneth A., and Jayshree Sarma. “Generation gaps revisited.” In Foundations of genetic algorithms, vol. 2, pp. 19-28. Elsevier, 1993.
- Parameters
offspring – list of created offpring, probably from pool()
parents – list of parents, usually the ones that offspring came from
k – how many elites from parents to keep?
key – optional key criteria for selecting; e.g., can be used to impose parsimony pressure
- Returns
surviving population, which will be offspring with offspring replaced by any superior parent elites
- leap_ec.ops.evaluate(next_individual: Iterator = '__no__default__') Iterator
Evaluate and returns the next individual in the pipeline
>>> from leap_ec.individual import Individual >>> from leap_ec.decoder import IdentityDecoder >>> from leap_ec.binary_rep.problems import MaxOnes >>> import numpy as np
We need to specify the decoder and problem so that evaluation is possible.
>>> genome = np.array([1, 1]) >>> ind = Individual(genome, decoder=IdentityDecoder(), problem=MaxOnes())
>>> evaluated_ind = next(evaluate(iter([ind])))
- Parameters
next_individual – iterator pointing to next individual to be evaluated
kwargs – contains optional context state to pass down the pipeline in context dictionaries
- Returns
the evaluated individual
- leap_ec.ops.grouped_evaluate(population: list = '__no__default__', max_individuals_per_chunk: int = None) list
Evaluate the population by sending groups of multiple individuals to a fitness function so they can be evaluated simultaneously.
This is useful, for example, as a way to evaluate individuals in parallel on a GPU.
- leap_ec.ops.insertion_selection(offspring: List = '__no__default__', parents: List = '__no__default__', key=None) List
do exclusive selection between offspring and parents
This is typically used for Ken De Jong’s EV algorithm for survival selection. Each offspring is deterministically selected and a random parent is selected; if the offspring wins, then it replaces the parent.
Note that we make a _copy_ of the parents and have the offspring compete with the parent copies so that users can optionally preserve the original parents. You may wish to do that, for example, if you want to analyze the composition of the original parents and the modified copy.
- Parameters
offspring – population to select from
parents – parents that are copied and which the copies are potentially updated with better offspring
key – optional key for determining max() by other criteria such as for parsimony pressure
- Returns
the updated parent population
- leap_ec.ops.iteriter_op(f)
This decorator wraps a function with runtime type checking to ensure that it always receives an iterator as its first argument, and that it returns an iterator.
We use this to make debugging operator pipelines easier in EAs: if you accidentally hook up, say an operator that outputs a list to an operator that expects an iterator, we’ll throw an exception that pinpoints the issue.
- Parameters
function (f) – the function to wrap
- leap_ec.ops.iterlist_op(f)
This decorator wraps a function with runtime type checking to ensure that it always receives an iterator as its first argument, and that it returns a list.
We use this to make debugging operator pipelines easier in EAs: if you accidentally hook up, say an operator that outputs a list to an operator that expects an iterator, we’ll throw an exception that pinpoints the issue.
- Parameters
function (f) – the function to wrap
- leap_ec.ops.listiter_op(f)
This decorator wraps a function with runtime type checking to ensure that it always receives a list as its first argument, and that it returns an iterator.
We use this to make debugging operator pipelines easier in EAs: if you accidentally hook up, say an operator that outputs an iterator to an operator that expects a list, we’ll throw an exception that pinpoints the issue.
- Parameters
function (f) – the function to wrap
- leap_ec.ops.listlist_op(f)
This decorator wraps a function with runtime type checking to ensure that it always receives a list as its first argument, and that it returns a list.
We use this to make debugging operator pipelines easier in EAs: if you accidentally hook up, say an operator that outputs an iterator to an operator that expects a list, we’ll throw an exception that pinpoints the issue.
- Parameters
function (f) – the function to wrap
- leap_ec.ops.migrate(topology, emigrant_selector, replacement_selector, migration_gap, customs_stamp=<function <lambda>>, metric=None, context={'leap': {'distrib': {'non_viable': 0}}})
A migration operator for use in island models.
This operator works with multi-population algorithms, and is thus meant to used with
leap_ec.algorithm.multi_population_ea.Specifically, it assumes that
the population argument passed into the returned function is a particular sub-population that we want to process “emigration” out of and “immigration” into,
the context state object contains an integer field context[‘leap’][‘generation’] indicating the current generation count of the algorithm, and
the context also contains a integer field context[‘leap’][‘current_subpopulation’] indicating the index of the subpopulation that is currently being processed in the overall collection of subpopulations (i.e. the one that population belongs to).
These assumptions are essentially what
leap_ec.algorithm.multi_population_eaimplements.>>> import networkx as nx >>> from leap_ec import ops, context >>> from leap_ec.data import test_population >>> pop0 = test_population[:] # Shallow copy >>> pop1 = test_population[:]
>>> op = migrate(topology=nx.complete_graph(2), ... emigrant_selector=ops.tournament_selection, ... replacement_selector=ops.random_selection, ... migration_gap=50) >>> context['leap']['generation'] = 0 >>> context['leap']['current_subpopulation'] = 0 >>> op(pop0) [Individual<...>(...), Individual<...>(...), Individual<...>(...), Individual<...>(...)]
>>> context['leap']['current_subpopulation'] = 1 >>> op(pop1) [Individual<...>(...), Individual<...>(...), Individual<...>(...), Individual<...>(...)]
This operator is a stateful closure: it maintains an internal list of all the out-going “emigrations” that occurred in the previous time step, so that it can process them as “immigrations” in the current time step.
- Parameters
topology – a networkx topology defining the connectivity among islands
emigrant_selector – a selection operator for choosing individuals to leave an island
replacement_selector – a selection operator choosing contestants that will be replaced by an incoming immigrant if the immigrant has higher fitness
migration_gap (int) – migration will occur regularly after every migration_gap evolutionary steps
customs_stamp – an optional function to transfrom an individual upon its arrival to a new island. This can be used, for example, to change the individual’s decoder or problem in a heterogeneous island model.
metric – an optional function of the form f(generation, immigrant_individual, contestant_indidivudal, success) for recording information about migration events.
context – the context object to check for EA state, such as the current generation number, and the ID of the subpopulation that is currently being processed.
- leap_ec.ops.migration_metric(stream, header: bool = True, notes: Optional[dict] = None)
Returns a function that can be used to record migration events.
The purpose of a migration metric is to record information about migrations that occur inside a migration operator. Because these events take place inside the operator (rather than across operators), they cannot be recorded by a LEAP pipeline probe.
In general, the interface for a migration metric function takes four parameters:
generation: the current generation
immigrant_ind: the individual that is attempting to migrate
contestant_ind: the individual that has been chosen to be replaced
success: True if the migration is successful, False otherwise
The metric included here records the fitness of both individuals and writes them (along with the generation and success values) to a CSV. You can write your own metric if you need to record other information (such as, say, genomes).
>>> import sys >>> from leap_ec import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> m = migration_metric(sys.stdout, ... header=True, ... notes={'run': 0, 'description': 'Test output'} ... ) run,description,generation,migrant_fitness,contestant_fitness,success
>>> ind1 = Individual(np.array([1, 1, 1]), problem=MaxOnes()) >>> f = ind1.evaluate() >>> contestant = Individual(np.array([0, 1, 1]), problem=MaxOnes()) >>> f = contestant.evaluate() >>> m(0, ind1, contestant, True) 0,Test output,0,3,2,True
- Parameters
stream – file object to write the CSV data to
header (bool) – a CSV header will be written if True
notes (dict) – a dict specifying additional constant-value columns to include in the CSV output
- leap_ec.ops.naive_cyclic_selection(population: List = '__no__default__', indices: List = None) Iterator
Deterministically returns individuals, and repeats the same test_sequence when exhausted.
This is “naive” because it doesn’t shuffle the population between complete tours to minimize bias.
>>> from leap_ec.individual import Individual >>> from leap_ec.ops import naive_cyclic_selection >>> import numpy as np
>>> pop = [Individual(np.array([0, 0])), ... Individual(np.array([0, 1]))]
>>> cyclic_selector = naive_cyclic_selection(pop)
- Parameters
population – from which to select
- Returns
the next selected individual
- leap_ec.ops.pool(next_individual: Iterator = '__no__default__', size: int = '__no__default__') List
‘Sink’ for creating size individuals from preceding pipeline source.
Allows for “pooling” individuals to be processed by next pipeline operator. Typically used to collect offspring from preceding set of selection and birth operators, but could also be used to, say, “pool” individuals to be passed to an EDA as a training set.
>>> from leap_ec.individual import Individual >>> from leap_ec.ops import naive_cyclic_selection >>> import numpy as np
>>> pop = [Individual(np.array([0, 0])), ... Individual(np.array([0, 1]))]
>>> cyclic_selector = naive_cyclic_selection(pop)
>>> pool = pool(cyclic_selector, 3)
print(pool) [Individual([0, 0], None, None), Individual([0, 1], None, None), Individual([0, 0], None, None)]
- Parameters
next_individual – generator for getting the next offspring
size – how many kids we want
- Returns
population of size offspring
- leap_ec.ops.proportional_selection(population: ~typing.List = '__no__default__', offset=0, exponent: int = 1, key=<function <lambda>>) Iterator
Returns an individual from a population in direct proportion to their fitness or another given metric.
To deal with negative fitness values use offset=’pop-min’ or set a custom offset. A ValueError is thrown if the result of adding offset to a fitness value results in a negative number. The value of an individual is calculated as follows
value = (fitness + offset)^exponent
- Parameters
population – the population to select from. Should be a list, not an iterator.
offset – the offset from zero. If negative fitness values are possible and the minimum is unknown use offest=’pop-min’ for an adaptive offset. Defaults to 0.
exponent (int) – the power to which fitness values are raised to. This can be tuned to increase or decrease selection pressure by creating larger or smaller differences between fitness values in the population. Defaults to 1.
key – a function that computes the metric used to compare individuals. Defaults to fitness.
- Returns
a random individual based on the proportion of the given metric in the population.
>>> from leap_ec import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.ops import proportional_selection >>> import numpy as np
>>> genome1 = np.array([0, 0, 0]) >>> genome2 = np.array([0, 0, 1]) >>> pop = [Individual(genome1, problem=MaxOnes()), ... Individual(genome2, problem=MaxOnes())] >>> pop = Individual.evaluate_population(pop) >>> selected = proportional_selection(pop)
- leap_ec.ops.random_bernoulli_vector(shape: Union[int, Tuple], p: float = 0.5) ndarray
Generates a random vector of Boolean balues from a Bernoulli process—that is, from a sequence of weighted coin flips.
We use this function throughout LEAP because its implementation was found to be much faster than, say, just calling np.random.choice([0, 1]).
>>> from leap_ec.ops import random_bernoulli_vector >>> random_bernoulli_vector(5, p=0.4) array([..., ..., ..., ..., ...])
- Parameters
shape – shape of the random vector—can be an integer or a tuple.
p – success probability of the bernoulli trials.
- Returns
boolean numpy array
- leap_ec.ops.random_selection(population: List = '__no__default__', indices=None) Iterator
return a uniformly randomly selected individual from the population
- Parameters
population – from which to select
- Returns
a uniformly selected individual
- leap_ec.ops.sus_selection(population: ~typing.List = '__no__default__', n=None, shuffle: bool = True, offset=0, exponent: int = 1, key=<function <lambda>>) Iterator
Returns an individual from a population in proportion to their fitness or another given metric using the stochastic universal sampling algorithm.
To deal with negative fitness values use offset=’pop-min’ or set a custom offset. A ValueError is thrown if the result of adding offset to a fitness value results in a negative number. The value of an individual is calculated as follows
value = (fitness + offset)^exponent
- Parameters
population – the population to select from. Should be a list, not an iterator.
n – the number of evenly spaced points to use in the algorithm. Default is None which uses len(population).
shuffle (bool) – if True, n points are resampled after one full pass over them. If False, selection repeats over the same n points. Defaults to True.
offset – the offset from zero. If negative fitness values are possible and the minimum is unknown use offset=’pop-min’ for an adaptive offset. Defaults to 0.
exponent (int) – the power to which fitness values are raised to. This can be tuned to increase or decrease selection pressure by creating larger or smaller differences between fitness values in the population. Defaults to 1.
key – a function that computes the metric used to compare individuals. Defaults to fitness.
- Returns
a random individual based on the proportion of the given metric in the population.
>>> from leap_ec import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.ops import sus_selection >>> import numpy as np
>>> genome1 = np.array([0, 0, 0]) >>> genome2 = np.array([1, 1, 1]) >>> pop = [Individual(genome1, problem=MaxOnes()), ... Individual(genome2, problem=MaxOnes())] >>> pop = Individual.evaluate_population(pop) >>> selected = sus_selection(pop)
- leap_ec.ops.tournament_selection(population: list = '__no__default__', k: int = 2, key=None, select_worst: bool = False, indices=None) Iterator
Returns an operator that selects the best individual from k individuals randomly selected from the given population.
Like other selection operators, this assumes that if one individual is “greater than” another, then it is “better than” the other. Whether this indicates maximization or minimization isn’t handled here: the Individual class determines the semantics of its “greater than” operator.
- Parameters
population – the population to select from. Should be a list, not an iterator.
k (int) – number of contestants in the tournament. k=2 does binary tournament selection, which approximates linear ranking selection in the expectation. Higher values of k yield greedier selection strategies—k=3, for instance, is equal to quadratic ranking selection in the expectation.
key – an optional function that computes keys to sort over. Defaults to None, in which case Individuals are compared directly.
select_worst (bool) – if True, select the worst individual from the tournament instead of the best.
indices (list) – an optional list that will be populated with the index of the selected individual.
- Returns
the best of k individuals drawn from population
>>> from leap_ec import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.ops import tournament_selection >>> import numpy as np
>>> pop = [Individual(np.array([0, 0, 0]), problem=MaxOnes()), ... Individual(np.array([0, 0, 1]), problem=MaxOnes())] >>> pop = Individual.evaluate_population(pop) >>> best = tournament_selection(pop)
- leap_ec.ops.truncation_selection(offspring: List = '__no__default__', size: int = '__no__default__', parents: List = None, key=None) List
return the size best individuals from the given population
This defaults to (mu, lambda) if parents is not given.
>>> from leap_ec.individual import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> from leap_ec.ops import truncation_selection >>> import numpy as np
>>> pop = [Individual(np.array([0, 0, 0]), problem=MaxOnes()), ... Individual(np.array([0, 0, 1]), problem=MaxOnes()), ... Individual(np.array([1, 1, 0]), problem=MaxOnes()), ... Individual(np.array([1, 1, 1]), problem=MaxOnes())]
We need to evaluate them to get their fitness to sort them for truncation.
>>> pop = Individual.evaluate_population(pop)
>>> truncated = truncation_selection(pop, 2)
TODO Do we want an optional context to over-ride the ‘parents’ parameter?
- Parameters
offspring – offspring to truncate down to a smaller population
size – is what to resize population to
second_population – is optional parent population to include with population for downsizing
- Returns
truncated population
leap_ec.parsimony module
Parsimony pressure functions.
These are intended to be used as key parameters for selection operators.
Provided are Koza-style parsimony pressure and lexicographic parsimony key functions.
- leap_ec.parsimony.koza_parsimony(ind='__no__default__', *, penalty='__no__default__')
Penalize fitness by genome length times a constant, in the style of Koza [Koz92].
>>> import toolz >>> from leap_ec.individual import Individual >>> from leap_ec.decoder import IdentityDecoder >>> from leap_ec.binary_rep.problems import MaxOnes >>> import leap_ec.ops as ops >>> import numpy as np >>> problem = MaxOnes() >>> pop = [Individual(np.array([0, 0, 0, 1, 1, 1]), problem=problem), ... Individual(np.array([0, 0]), problem=problem), ... Individual(np.array([1, 1]), problem=problem), ... Individual(np.array([1, 1, 1]), problem=problem)] >>> pop = Individual.evaluate_population(pop) >>> best, = ops.truncation_selection(pop, size=1) >>> print(best.genome, best.fitness) [0 0 0 1 1 1] 3
>>> best, = ops.truncation_selection(pop, size=1, key=koza_parsimony(penalty=.5)) >>> print(best.genome, best.fitness) [1 1 1] 3
\[f_p(x) = f(x) - cl(x)\]Where f(x) is original fitness, c is a penalty constant, and l(x) is the genome length.
- Parameters
ind – to be compared
penalty – for denoting penalty strength
- Returns
altered comparison criteria
- leap_ec.parsimony.lexical_parsimony(ind)
If two fitnesses are the same, break the tie with the smallest genome
This implements Lexicographical Parsimony Pressure [LP02], which is essentially where if the fitnesses of two individuals are close, then break the tie with the smallest genome.
>>> import toolz >>> from leap_ec.individual import Individual >>> from leap_ec.binary_rep.problems import MaxOnes >>> import leap_ec.ops as ops >>> import numpy as np >>> problem = MaxOnes() >>> pop = [Individual(np.array([0, 0, 0, 1, 1, 1]), problem=problem), ... Individual(np.array([0, 0]), problem=problem), ... Individual(np.array([1, 1]), problem=problem), ... Individual(np.array([1, 1, 1]), problem=problem)] >>> pop = Individual.evaluate_population(pop) >>> best, = ops.truncation_selection(pop, size=1) >>> print(best.genome, best.fitness) [0 0 0 1 1 1] 3
>>> best, = ops.truncation_selection(pop, size=1, key=lexical_parsimony) >>> print(best.genome, best.fitness) [1 1 1] 3
- Parameters
ind – to be compared
- Returns
altered comparison criteria
leap_ec.probe module
Probes are pipeline operators to instrument state that passes through the pipeline such as populations or individuals.
- class leap_ec.probe.AttributesCSVProbe(attributes=(), stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>, do_dataframe=False, best_only=False, header=True, do_fitness=False, do_genome=False, notes=None, extra_metrics=None, job=None, numpy_as_list=True, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
OperatorAn operator that records the specified attributes for all the individuals (or just the best individual) in population in CSV-format to the specified stream and/or to a DataFrame.
- Parameters
attributes – list of attribute names to record, as found in the individuals’ attributes field
stream – a file object to write the CSV rows to (defaults to sys.stdout). Can be None if you only want a DataFrame
do_dataframe (bool) – if True, data will be collected in memory as a Pandas DataFrame, which can be retrieved by calling the dataframe property after (or during) the algorithm run. Defaults to False, since this can consume a lot of memory for long-running algorithms.
best_only (bool) – if True, attributes will only be recorded for the best-fitness individual; otherwise a row is recorded for every individual in the population
header (bool) – if True (the default), a CSV header is printed as the first row with the column names
do_fitness (bool) – if True, the individuals’ fitness is included as one of the columns
do_genomes (bool) – if True, the individuals’ genome is included as one of the columns
notes (str) – a dict of optional constant-value columns to include in all rows (ex. to identify and experiment or parameters)
extra_metrics – a dict of ‘column_name’: function pairs, to compute optional extra columns. The functions take a the population as input as a list of individuals, and their return value is printed in the column.
job (int) – a job ID that will be included as a constant-value column in all rows (ex. typically an integer, indicating the ith run out of many)
numpy_as_list (bool) – if True, numpy arrays will be first converted to a python list before printing. This is intended for large genomes and multiobjective fitnesses, where large numpy arrays would be split across multiple csv rows by the default formatter.
context – the algorithm context we use to read the current generation from (so we can write it to a column)
Individuals contain some build-in attributes (namely fitness, genome), and also a dict of additional custom attributes called, well, attributes. This class allows you to log all of the above.
Most often, you will want to record only the best individual in the population at each step, and you’ll just want to know its fitness and genome. You can do this with this class’s boolean flags. For example, here’s how you’d record the best individual’s fitness and genome to a dataframe:
>>> from leap_ec.global_vars import context >>> from leap_ec.data import test_population >>> probe = AttributesCSVProbe(do_dataframe=True, best_only=True, ... do_fitness=True, do_genome=True) >>> context['leap']['generation'] = 100 >>> probe(test_population) == test_population True
You can retrieve the result programatically from the dataframe property:
>>> probe.dataframe step fitness genome 0 100 4 [0, 1, 1, 1, 1]
By default, the results are also written to sys.stdout. You can pass any file object you like into the stream parameter.
Another common use of this task is to record custom attributes that are stored on individuals in certain kinds of experiments. Here’s how you would record the values of ind.foo and ind.bar for every individual in the population. We write to a stream object this time to demonstrate how to use the probe without a dataframe:
>>> import io >>> stream = io.StringIO() >>> probe = AttributesCSVProbe(attributes=['foo', 'bar'], stream=stream) >>> context['leap']['generation'] = 100 >>> r = probe(test_population) >>> print(stream.getvalue()) step,foo,bar 100,GREEN,Colorless 100,15,green 100,BLUE,ideas 100,72.81,sleep
- property dataframe
Property for retrieving a Pandas DataFrame representation of the collected data.
- get_row_dict(ind)
Compute a full row of data from a given individual.
- class leap_ec.probe.BestSoFarIterProbe(stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>, header=True, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
Operator- This probe takes an iterator as input and will track the
best-so-far (BSF) individual in the all the individuals it sees.
Insert an object of this class into a pipeline to have it track the the best individual it sees so far. It will write the current best individual for each __call__ invocation to a given stream in CSV format.
Like many operators, this operator checks the context object to retrieve the current generation number for output purposes.
>>> from leap_ec import context, data >>> from leap_ec import probe >>> pop = data.test_population >>> context['leap']['generation'] = 12
The probe will write its output to the provided stream (default is stdout, but we illustrate here with a StringIO stream):
>>> import io >>> stream = io.StringIO() >>> probe = BestSoFarIterProbe(stream=stream) >>> bsf_output_iter = probe(iter(pop)) >>> x = next(bsf_output_iter) >>> x = next(bsf_output_iter) >>> x = next(bsf_output_iter) >>> print(stream.getvalue()) step,bsf 12,... 12,... 12,...
- class leap_ec.probe.BestSoFarProbe(stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>, header=True, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
Operator- This probe takes an list of individuals as input and will track the
best-so-far (BSF) individual across all the population it has seen.
Insert an object of this class into a pipeline to have it track the the best individual it sees so far. It will write the current best individual for each __call__ invocation to a given stream in CSV format.
Like many operators, this operator checks the context object to retrieve the current generation number for output purposes.
>>> from leap_ec import context, data >>> from leap_ec import probe >>> pop = data.test_population >>> context['leap']['generation'] = 12
The probe will write its output to the provided stream (default is stdout, but we illustrate here with a StringIO stream):
>>> import io >>> stream = io.StringIO() >>> probe = BestSoFarProbe(stream=stream) >>> new_pop = probe(pop) >>> print(stream.getvalue()) step,bsf 12,4
This operator does not change the state of the population: >>> new_pop == pop True
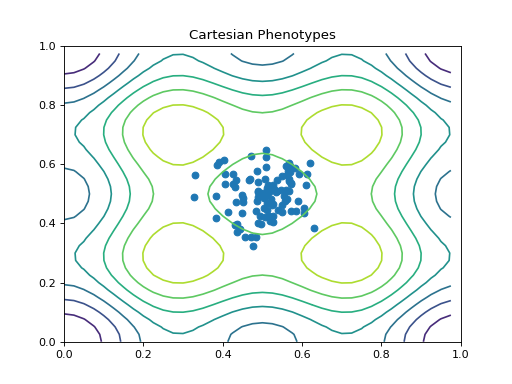
- class leap_ec.probe.CartesianPhenotypePlotProbe(ax=None, xlim=(-5.12, 5.12), ylim=(-5.12, 5.12), contours=None, granularity=None, title='Cartesian Phenotypes', modulo=1, context={'leap': {'distrib': {'non_viable': 0}}}, pad=())
Bases:
objectMeasure and plot a scatterplot of the populations’ location in a 2-D phenotype space.
- Parameters
ax (Axes) – Matplotlib axes to plot to (if None, a new figure will be created).
xlim ((float, float)) – Bounds of the horizontal axis.
ylim ((float, float)) – Bounds of the vertical axis.
contours (Problem) – a problem defining a 2-D fitness function (this will be used to draw fitness contours in the background of the scatterplot).
granularity (float) – (Optional) spacing of the grid to sample points along while drawing the fitness contours. If none is given, then the granularity will default to 1/50th of the range of the function’s bounds attribute.
modulo (int) – take and plot a measurement every modulo steps ( default 1).
pad – A list of extra gene values, used to fill in the hidden dimensions with contants while drawing fitness contours.
Attach this probe to matplotlib
Axesand then insert it into an EA’s operator pipeline to get a live phenotype plot that updates every modulo steps.>>> import matplotlib.pyplot as plt >>> from leap_ec.probe import CartesianPhenotypePlotProbe >>> from leap_ec.representation import Representation
>>> from leap_ec.individual import Individual >>> from leap_ec.algorithm import generational_ea
>>> from leap_ec import ops >>> from leap_ec.decoder import IdentityDecoder >>> from leap_ec.real_rep.problems import CosineFamilyProblem >>> from leap_ec.real_rep.initializers import create_real_vector >>> from leap_ec.real_rep.ops import mutate_gaussian
>>> # The fitness landscape >>> problem = CosineFamilyProblem(alpha=1.0, global_optima_counts=[2, 2], local_optima_counts=[2, 2])
>>> # If no axis is provided, a new figure will be created for the probe to write to >>> trajectory_probe = CartesianPhenotypePlotProbe(contours=problem, ... xlim=(0, 1), ylim=(0, 1), ... granularity=0.025)
>>> # Create an algorithm that contains the probe in the operator pipeline
>>> pop_size = 100 >>> ea = generational_ea(max_generations=20, pop_size=pop_size, ... problem=problem, ... ... representation=Representation( ... individual_cls=Individual, ... initialize=create_real_vector(bounds=[[0.4, 0.6]] * 2), ... decoder=IdentityDecoder() ... ), ... ... pipeline=[ ... trajectory_probe, # Insert the probe into the pipeline like so ... ops.tournament_selection, ... ops.clone, ... mutate_gaussian(std=0.05, expected_num_mutations='isotropic', bounds=(0, 1)), ... ops.evaluate, ... ops.pool(size=pop_size) ... ]) >>> result = list(ea);
(Source code, png, hires.png, pdf)
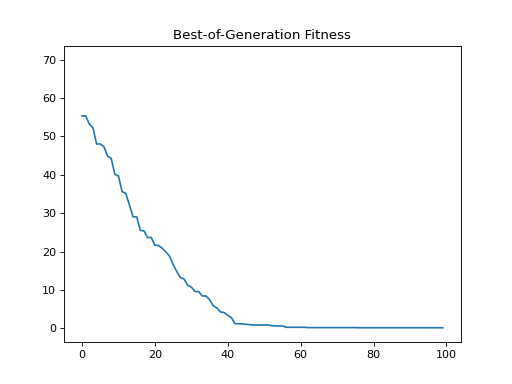
- class leap_ec.probe.FitnessPlotProbe(ax=None, xlim=None, ylim=None, modulo=1, title='Best-of-Generation Fitness', x_axis_value=None, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
PopulationMetricsPlotProbeMeasure and plot a population’s fitness trajectory.
- Parameters
ax (Axes) – Matplotlib axes to plot to (if None, a new figure will be created).
xlim ((float, float)) – Bounds of the horizontal axis.
ylim ((float, float)) – Bounds of the vertical axis.
modulo (int) – take and plot a measurement every modulo steps ( default 1).
title – title to print on the plot
x_axis_value – optional function to define what value gets plotted on the x axis. Defaults to pulling the ‘generation’ value out of the default context object.
context – set a context object to query for the current generation. Defaults to the standard leap_ec.context object.
Attach this probe to matplotlib
Axesand then insert it into an EA’s operator pipeline.>>> import matplotlib.pyplot as plt >>> from leap_ec.probe import FitnessPlotProbe >>> from leap_ec.representation import Representation
>>> f = plt.figure() # Setup a figure to plot to >>> plot_probe = FitnessPlotProbe(ylim=(0, 70), ax=plt.gca())
>>> # Create an algorithm that contains the probe in the operator pipeline >>> from leap_ec.individual import Individual >>> from leap_ec.decoder import IdentityDecoder >>> from leap_ec import ops >>> from leap_ec.real_rep.problems import SpheroidProblem >>> from leap_ec.real_rep.ops import mutate_gaussian >>> from leap_ec.real_rep.initializers import create_real_vector
>>> from leap_ec.algorithm import generational_ea
>>> l = 10 >>> pop_size = 10 >>> ea = generational_ea(max_generations=100, pop_size=pop_size, ... problem=SpheroidProblem(maximize=False), ... ... representation=Representation( ... individual_cls=Individual, ... decoder=IdentityDecoder(), ... initialize=create_real_vector(bounds=[[-5.12, 5.12]] * l) ... ), ... ... pipeline=[ ... plot_probe, # Insert the probe into the pipeline like so ... ops.tournament_selection, ... ops.clone, ... mutate_gaussian(std=0.2, expected_num_mutations='isotropic'), ... ops.evaluate, ... ops.pool(size=pop_size) ... ]) >>> result = list(ea);
(Source code, png, hires.png, pdf)
To get a live-updated plot that words like a real-time video of the EA’s progress, use this probe in conjunction with the %matplotlib notebook magic for Jupyter Notebook (as opposed to %matplotlib inline, which only allows static plots).
- class leap_ec.probe.FitnessStatsCSVProbe(stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>, header=True, extra_metrics=None, comment=None, job: ~typing.Optional[str] = None, notes: ~typing.Optional[~typing.Dict] = None, modulo: int = 1, numpy_as_list=True, context: ~typing.Dict = {'leap': {'distrib': {'non_viable': 0}}})
Bases:
OperatorA probe that records basic fitness statistics for a population to a text stream in CSV format.
This is meant to capture the “bread and butter” values you’ll typically want to see in any population-based optimization experiment. If you want additional columns with custom values, you can pass in a dict of notes with constant values or extra_metrics with functions to compute them.
- Parameters
stream – the file object to write to (defaults to sys.stdout)
header – whether to print column names in the first line
extra_metrics – a dict of ‘column_name’: function pairs, to compute optional extra columns. The functions take a the population as input as a list of individuals, and their return value is printed in the column.
job – optional constant job ID, which will be printed as the first column
notes (str) – a dict of optional constant-value columns to include in all rows (ex. to identify and experiment or parameters)
numpy_as_list (bool) – if True, numpy arrays will be first converted to a python list before printing. This is intended for multiobjective fitnesses, where large numpy arrays are normally split across csv rows with the default formatter.
context – a LEAP context object, used to retrieve the current generation from the EA state (i.e. from context[‘leap’][‘generation’])
In this example, we’ll set up two three inputs for the probe: an output stream, the generation number, and a population.
We use a StringIO stream to print the results here, but in practice you often want to use sys.stdout (the default) or a file object:
>>> import io >>> stream = io.StringIO()
The probe also relies on LEAP’s algorithm context to determine the generation number:
>>> from leap_ec.global_vars import context >>> context['leap']['generation'] = 100
Here’s how we’d compute fitness statistics for a test population. The population is unmodified:
>>> from leap_ec.data import test_population >>> probe = FitnessStatsCSVProbe(stream=stream, job=15, notes={'description': 'just a test'}) >>> probe(test_population) == test_population True
and the output has the following columns: >>> print(stream.getvalue()) job, description, step, bsf, mean_fitness, std_fitness, min_fitness, max_fitness 15, just a test, 100, 4, 2.5, 1.11803…, 1, 4 <BLANKLINE>
To add custom columns, use the extra_metrics dict. For example, here’s a function that computes the median fitness value of a population:
>>> import numpy as np >>> median = lambda p: np.median([ ind.fitness for ind in p ])
We can include it in the fitness stats report like so:
>>> stream = io.StringIO() >>> extras_probe = FitnessStatsCSVProbe(stream=stream, job="15", extra_metrics={'median_fitness': median}) >>> extras_probe(test_population) == test_population True
>>> print(stream.getvalue()) job, step, bsf, mean_fitness, std_fitness, min_fitness, max_fitness, median_fitness 15, 100, 4, 2.5, 1.11803..., 1, 4, 2.5
- comment_character = '#'
- default_metric_cols = ('bsf', 'mean_fitness', 'std_fitness', 'min_fitness', 'max_fitness')
- time_col = 'step'
- write_comment(stream)
- write_header(stream)
- class leap_ec.probe.HeatMapPhenotypeProbe(ax=None, title='HeatMap of Phenotypes', modulo=1, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
object
- class leap_ec.probe.HistPhenotypePlotProbe(ax=None, title='Histogram of Phenotypes', modulo=1, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
objectA visualization probe that uses matplotlib to show a live histogram of the population’s phenotypes.
This typically makes the most since for 1-dimensional genotypes.
- class leap_ec.probe.PopulationMetricsPlotProbe(ax=None, metrics=None, xlim=None, ylim=None, modulo=1, title='Population Metrics', x_axis_value=None, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
object- reset()
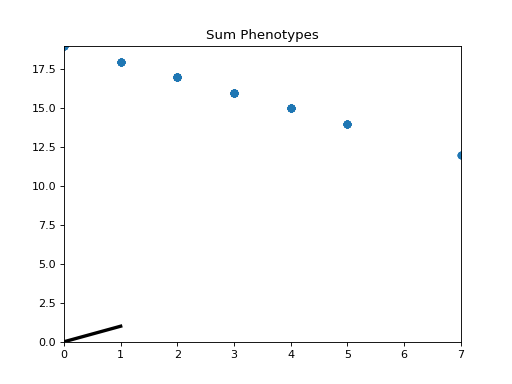
- class leap_ec.probe.SumPhenotypePlotProbe(ax=None, xlim=(-5.12, 5.12), ylim=(-5.12, 5.12), problem=None, granularity=1, title='Sum Phenotypes', modulo=1, context={'leap': {'distrib': {'non_viable': 0}}})
Bases:
objectPlot the population’s location on a fitness landscape that is defined over the sum of a vector phenotype’s elements. This is useful for visualizing OneMax functions and similar functions that can be understood in terms of a graph with “the number of ones” along the x axis.
- Parameters
ax (Axes) – Matplotlib axes to plot to (if None, a new figure will be created).
xlim ((float, float)) – Bounds of the horizontal axis.
ylim ((float, float)) – Bounds of the vertical axis.
problem (Problem) – a problem that will be used to draw a fitness curve.
granularity (float) – (Optional) spacing of the grid to sample points along while drawing the fitness contours. If none is given, then the granularity will default to 1.0.
modulo (int) – take and plot a measurement every modulo steps ( default 1).
Attach this probe to matplotlib
Axesand then insert it into an EA’s operator pipeline to get a live phenotype plot that updates every modulo steps.>>> import matplotlib.pyplot as plt >>> from leap_ec.probe import SumPhenotypePlotProbe >>> from leap_ec.representation import Representation
>>> from leap_ec.individual import Individual >>> from leap_ec.algorithm import generational_ea
>>> from leap_ec import ops >>> from leap_ec.binary_rep.problems import DeceptiveTrap >>> from leap_ec.binary_rep.initializers import create_binary_sequence >>> from leap_ec.binary_rep.ops import mutate_bitflip
>>> # The fitness landscape >>> problem = DeceptiveTrap()
>>> # If no axis is provided, a new figure will be created for the probe to write to >>> dimensions = 20 >>> trajectory_probe = SumPhenotypePlotProbe(problem=problem, ... xlim=(0, dimensions), ylim=(0, dimensions))
>>> # Create an algorithm that contains the probe in the operator pipeline
>>> pop_size = 100 >>> ea = generational_ea(max_generations=20, pop_size=pop_size, ... problem=problem, ... ... representation=Representation( ... individual_cls=Individual, ... initialize=create_binary_sequence(length=dimensions) ... ), ... ... pipeline=[ ... trajectory_probe, # Insert the probe into the pipeline like so ... ops.tournament_selection, ... ops.clone, ... mutate_bitflip(expected_num_mutations=1), ... ops.evaluate, ... ops.pool(size=pop_size) ... ]) >>> result = list(ea);
(Source code, png, hires.png, pdf)
- leap_ec.probe.best_of_gen(population)
Syntactic sugar to select the best individual in a population.
- Parameters
population – a list of individuals
context – optional dict of auxiliary state (ignored)
>>> from leap_ec.data import test_population >>> print(best_of_gen(test_population)) Individual<...> with fitness 4
- leap_ec.probe.num_fixated_metric(population: list)
Computes the genetic diversity of the population by counting the number of variables in the genome that have zero variance.
This is a so-called “column-wise” metric, in the sense that it considers each element of the solution vectors independently.
- leap_ec.probe.pairwise_squared_distance_metric(population: list)
Computes the genetic diversity of a population by considering the sum of squared Euclidean distances between individual genomes.
We compute this in \(O(n)\) by writing the sum in terms of distance from the population centroid \(c\):
\[\mathcal{D}(\text{population}) = \sum_{i=1}^n \sum_{j=1}^n \| x_i - x_j \|^2 = 2n \sum_{i=1}^n \| x_i - c \|^2\]
- leap_ec.probe.print_individual(next_individual: ~typing.Iterator = '__no__default__', prefix='', numpy_as_list=False, stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>) Iterator
Just echoes the individual from within the pipeline
Uses next_individual.__str__
- Parameters
next_individual – iterator for next individual to be printed
prefix – prefix appended to the start of the line
numpy_as_list – If True, numpy arrays are converted to lists before printing
stream – File object passed to print
- Returns
the same individual, unchanged
- leap_ec.probe.print_population(population, generation, numpy_as_list=False)
Convenience function for pretty printing a population that’s associated with a given generation
- Parameters
population – The population of individuals to be printed
generation – The generation of the population
numpy_as_list – If True, numpy arrays are converted to lists before printing
- Returns
None
- leap_ec.probe.print_probe(population='__no__default__', probe='__no__default__', stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>, prefix='')
pipeline operator for printing the given population
This is really a wrapper around probe that, itself, gets passed te entire population.
The optional prefix is used to tag the output. For example, you may want to print ‘before’ to indicate that the population is before an operator is applied.
- Parameters
population – to be printed
probe – secondary probe that gets the poplation as input and for which the output is passed to stream
stream – to write output
prefix – optional string prefix to prepend to output
- Returns
population
- leap_ec.probe.sum_of_variances_metric(population: list)
Computes the genetic diversity of a population by considering the sum of the variances of each variable in the genome.
\[\mathcal{D}(\text{population}) = \sum_{i=1}^L \mathbb{E}_{j \in P}\left[ x_j[i] - \mathbb{E}[x_j[i]] \right]\]This is a so-called “column-wise” metric, in the sense that it considers each element of the solution vectors independently.
leap_ec.problem module
Defines the abstract-base classes Problem, ScalarProblem, and FunctionProblem.
- class leap_ec.problem.AlternatingProblem(problems, modulo, context={'leap': {'distrib': {'non_viable': 0}, 'generation': 20}})
Bases:
Problem- equivalent(first_fitness, second_fitness)
- evaluate(phenome)
Evaluate the given phenome.
Practitioners must over-ride this member function.
Note that by default the individual comparison operators assume a maximization problem; if this is a minimization problem, then just negate the value when returning the fitness.
- Parameters
phenome – the phenome to evaluate (this will not be modified)
- Returns
the fitness value
- get_current_problem()
- worse_than(first_fitness, second_fitness)
- class leap_ec.problem.AverageFitnessProblem(wrapped_problem, n: int)
Bases:
ProblemProblem wrapper that copies each genome n times, evaluates them, and averages the results back together to produce a mean-fitness estimate.
This is a common strategy for approaching noisy fitness functions, to make it easier for an optimization algorithm to follow a gradient.
>>> from leap_ec.real_rep.problems import NoisyQuarticProblem >>> p = AverageFitnessProblem( ... wrapped_problem = NoisyQuarticProblem(), ... n = 20) >>> x = [ 1, 1, 1, 1 ] >>> y = p.evaluate(x) >>> print(f"Fitness: {y}") # The mean of this will be approximately 10 Fitness: ...
- equivalent(first_fitness, second_fitness)
- evaluate(phenome)
Evaluates the wrapped function n times sequentially and returns the mean.
- evaluate_multiple(phenomes: list)
Evaluate a collections of phenomes by creating n jobs for each phenome, sending all the jobs to the wrapped evaluate_multiple() function, and then averaging the n results for each phenome into a list of results.
- worse_than(first_fitness, second_fitness)
- class leap_ec.problem.ConstantProblem(maximize=False, c=1.0)
Bases:
ScalarProblemA flat landscape, where all phenotypes have the same fitness.
This is sometimes useful for sanity checks or as a control in certain kinds of research.
\[f(\vec{x}) = c\]- Parameters
c (float) – the fitness value to return for any input.
from leap_ec.problem import ConstantProblem from leap_ec.real_rep.problems import plot_2d_problem bounds = ConstantProblem.bounds plot_2d_problem(ConstantProblem(), xlim=bounds, ylim=bounds, granularity=0.025)
(Source code, png, hires.png, pdf)
- bounds = (-1.0, 1.0)
- evaluate(phenome, *args, **kwargs)
Return a contant value for any input phenome:
>>> phenome = [0.5, 0.8, 1.5] >>> ConstantProblem().evaluate(phenome) 1.0
>>> ConstantProblem(c=500.0).evaluate('foo bar') 500.0
- Parameters
phenome – phenome to be evaluated
- Returns
1.0, or the constant defined in the constructor
- class leap_ec.problem.CooperativeProblem(wrapped_problem, num_trials: int, collaborator_selector, combined_decoder: ~leap_ec.decoder.Decoder = IdentityDecoder(), log_stream=None, combine_genomes=<function CooperativeProblem.<lambda>>, context={'leap': {'distrib': {'non_viable': 0}, 'generation': 20}})
Bases:
ProblemA Problem that implements cooperative coevolution. This provides a fitness function that takes partial solutions as input (i.e. from one of the subpopulations of the cooperative algorithm), and evaluates their fitness by combining them with other individuals in the population.
You can think of a CooperativeProblem as defining a fitness function for a subpopulation in a multi-population model, where the fitness function that is computed is itself a function of the state of the other subpopulations:
..math
mbox{fitness} = f_{p_i}(vec{mathbf{x}}, mathcal{P} \ p_i)
This class works by wrapping another fitness function, which is defined over complete solutions, and by taking a selection operator (which is used to select “collaborators” from other subpopulations to form complete solutions):
>>> from leap_ec import ops >>> from leap_ec.real_rep.problems import SpheroidProblem >>> complete_problem = SpheroidProblem() >>> problem = CooperativeProblem( ... wrapped_problem = SpheroidProblem(), ... num_trials = 3, ... collaborator_selector = ops.random_selection)
- equivalent(first_fitness, second_fitness)
- evaluate(phenome, *args, **kwargs)
Evaluate the given phenome.
Practitioners must over-ride this member function.
Note that by default the individual comparison operators assume a maximization problem; if this is a minimization problem, then just negate the value when returning the fitness.
- Parameters
phenome – the phenome to evaluate (this will not be modified)
- Returns
the fitness value
- evaluate_multiple(phenomes, individuals)
Evaluate multiple phenomes all at once, returning a list of fitness values.
By default this just calls self.evaluate() multiple times. Override this if you need to, say, send a group of individuals off to parallel
- worse_than(first_fitness, second_fitness)
- class leap_ec.problem.ExternalProcessProblem(command: str, maximize: bool, args: Optional[list] = None)
Bases:
ScalarProblemEvaluate individuals by launching an external program, writing phenomes to its stdin as CSV rows, and reading back fitness values from its stdout.
Assumes that individuals are represented with list phenomes with elements that can be cast to strings.
- evaluate(phenome)
Evaluate the given phenome.
Practitioners must over-ride this member function.
Note that by default the individual comparison operators assume a maximization problem; if this is a minimization problem, then just negate the value when returning the fitness.
- Parameters
phenome – the phenome to evaluate (this will not be modified)
- Returns
the fitness value
- evaluate_multiple(phenomes, *args, **kwargs)
Evaluate multiple phenomes all at once, returning a list of fitness values.
By default this just calls self.evaluate() multiple times. Override this if you need to, say, send a group of individuals off to parallel
- class leap_ec.problem.FitnessOffsetProblem(problem, fitness_offset, maximize=None)
Bases:
ScalarProblemTakes an existing function and adds a constant value to it output.
\[f'(\mathbf{x}) = f(\mathbf{x}) + c\]- Parameters
problem – the original problem to wrape
fitness_offset (float) – the scalar constant to add
- evaluate(phenome)
Evaluates the phenome’s fitness in the wrapped function, then adds the constant.
For example, here the original fitness function returns 5.0, but we subtract 3.5 from it so that it yields 1.5.
>>> original = ConstantProblem(c=5.0) >>> problem = FitnessOffsetProblem(original, fitness_offset=-3.5) >>> problem.evaluate([0, 1, 2]) 1.5
- class leap_ec.problem.FunctionProblem(fitness_function, maximize)
Bases:
ScalarProblemA convenience wrapper that takes a vanilla function that returns scalar fitness values and makes it usable as an objective function.
- evaluate(phenome, *args, **kwargs)
Evaluate the given phenome.
Practitioners must over-ride this member function.
Note that by default the individual comparison operators assume a maximization problem; if this is a minimization problem, then just negate the value when returning the fitness.
- Parameters
phenome – the phenome to evaluate (this will not be modified)
- Returns
the fitness value
- class leap_ec.problem.Problem
Bases:
ABCAbstract Base Class used to define problem definitions.
A Problem is in charge of two major parts of an EA’s behavior:
Fitness evaluation (the evaluate() method)
Fitness comparision (the worse_than() and equivalent() methods)
- abstract equivalent(first_fitness, second_fitness)
- abstract evaluate(phenome, *args, **kwargs)
Evaluate the given phenome.
Practitioners must over-ride this member function.
Note that by default the individual comparison operators assume a maximization problem; if this is a minimization problem, then just negate the value when returning the fitness.
- Parameters
phenome – the phenome to evaluate (this will not be modified)
- Returns
the fitness value
- evaluate_multiple(phenomes)
Evaluate multiple phenomes all at once, returning a list of fitness values.
By default this just calls self.evaluate() multiple times. Override this if you need to, say, send a group of individuals off to parallel
- abstract worse_than(first_fitness, second_fitness)
- class leap_ec.problem.ScalarProblem(maximize)
Bases:
ProblemA problem that compares individuals based on their scalar fitness values.
Inherit from this class and implement the evaluate() method to implement an objective function that returns a single real-valued fitness value.
- equivalent(first_fitness, second_fitness)
Used in Individual.__eq__().
By default returns first.fitness== second.fitness. Please over-ride if this does not hold for your problem.
- Returns
true if the first individual is equal to the second
- worse_than(first_fitness, second_fitness)
Used in Individual.__lt__().
By default returns first_fitness < second_fitness if a maximization problem, else first_fitness > second_fitness if a minimization problem. Please over-ride if this does not hold for your problem.
- Returns
true if the first individual is less fit than the second
- leap_ec.problem.concat_combine(collaborators)
Combine a list of individuals by concatenating their genomes.
This is a convenience function intended for use with CooperativeProblem.
leap_ec.representation module
A Representation is a simple data structure that wraps the components needed to define, initialize, and decode individuals.
This just serves as some syntactic sugar when we are specifying algorithms—so that representation-related components are grouped together and clearly labeled Representation.
- class leap_ec.representation.Representation(initialize, decoder=IdentityDecoder(), individual_cls=<class 'leap_ec.individual.Individual'>)
Bases:
objectSyntactic sugar for some of the monolithic functions that conveniently combines a decoder, initializer, and an Individual class since those always work in tandem, but can still be loosely coupled.
- create_individual(problem)
Make a single individual.
- create_population(pop_size, problem)
make a new population
- Parameters
pop_size – how many individuals should be in the population
problem – to be solved
- Returns
a population of individual_cls individuals
leap_ec.simple module
Provides a very high-level convenience function for a very general EA, ea_solve().
- leap_ec.simple.ea_solve(function, bounds, generations=100, pop_size=2, mutation_std=1.0, maximize=False, viz=False, viz_ylim=(0, 1), hard_bounds=True, dask_client=None, stream=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='UTF-8'>)
Provides a simple, top-level interfact that optimizes a real-valued function using a simple generational EA.
- Parameters
function – the function to optimize; should take lists of real numbers as input and return a float fitness value
bounds ([(float, float)]) – a list of (min, max) bounds to define the search space
generations (int) – the number of generations to run for
pop_size (int) – the population size
mutation_std (float) – the width of the mutation distribution
maximize (bool) – whether to maximize the function (else minimize)
viz (bool) – whether to display a live best-of-generation plot
hard_bounds (bool) – if True, bounds are enforced at all times during evolution; otherwise they are only used to initialize the population.
viz_ylim ((float, float)) – initial bounds to use of the plots vertical axis
dask_client – is optional dask Client to enable parallel evaluations
stream – a stream to write best-so-far values to (defaults to stdout)
The basic call includes instrumentation that prints the best-so-far fitness value of each generation to stdout:
>>> import io >>> from leap_ec.simple import ea_solve >>> stream = io.StringIO() >>> ea_solve(sum, bounds=[(0, 1)]*5, stream=stream) array([..., ..., ..., ..., ...])
The stream captures the best-so-far individual at each iteration of the algorithm: >>> print(stream.getvalue()) # doctest: +ELLIPSIS, +NORMALIZE_WHITESPACE step,bsf 0,… 1,… 2,… … 99,… <BLANKLINE>
When viz=True, a live BSF plot will also display:
>>> ea_solve(sum, bounds=[(0, 1)]*5, viz=True) array([..., ..., ..., ..., ...])
(Source code, png, hires.png, pdf)
leap_ec.statistical_helpers module
Helpers for testing the output of stochastic functions.
- leap_ec.statistical_helpers.collect_distribution(function, samples: int)
Count the number of times the given function returns each output value.
- leap_ec.statistical_helpers.equals_gaussian(observed_samples, reference_mean: float, reference_std: float, num_reference_observations: int, p: float) bool
A convenience function for computing a t-test for equality of two independent samples, using samples from one and test statistics from the other.
Assumes equal variance samples.
>>> import numpy as np >>> observed = np.random.normal(15, 1, size=1000) >>> equals_gaussian(observed, 15, 1, 1000, p=0.05) True
- leap_ec.statistical_helpers.equals_uniform(observed_distribution: Dict, p: float) bool
Use a $chi^2$ test to determine whether the observed distribution is uniform.
This offers convenience over stochastic_equals(), because the expected distribution doesn’t have to be manually specified.
For example, we do not reject the hypothesis that [5060, 4940] comes from a uniform distribution:
>>> observed = { 0: 5060, 1: 4940 } >>> equals_uniform(observed, p=0.01) True
The keys are arbitrary, so we can use them to clearly express what we are testing:
>>> observed = { 'Left': 101, 'Right': 100, 'Up': 99, 'Down': 100 } >>> equals_uniform(observed, p=0.01) True
- leap_ec.statistical_helpers.stochastic_chisquare(expected_distribution, distribution)
Use a $chi^2$ distribution to compute a p-value for the probability of rejecting the hypothesis that the given distribution matches the expected distribution.
The null hypothesis here is that the distributions are equal.
This takes two dictionaries of values:
>>> expected_distribution = { 1: 10, 2: 10, 3: 10, 4: 10, 5: 10, 6: 10} >>> distribution = { 1: 5, 2: 8, 3: 9, 4: 8, 5: 10, 6: 20} >>> stochastic_chisquare(expected_distribution, distribution) 0.01990...
The p-value is low, because the samples are quite different, so the “probability of seeing that big a difference or greater if the two distributions are equal” is low.
Very similar samples, by contrast, yield high p values:
>>> expected_distribution = { 1: 10, 2: 10, 3: 10, 4: 10, 5: 10, 6: 10} >>> distribution = { 1: 10, 2: 10, 3: 10, 4: 10, 5: 10, 6: 10} >>> stochastic_chisquare(expected_distribution, distribution) 1.0
- leap_ec.statistical_helpers.stochastic_equals(expected_distribution: Dict, observed_distribution: Dict, p: float) bool
Use a $chi^2$ test to determine whether two discrete distributions are equal. Specifically, this returns false if we reject the hypothesis that they are equal at the given p-value.
Lower p-value thresholds make the test more conservative, in the sense that it will assume the distributions are equal unless there is very good evidence to the contrary. We typically want to use low p-value thresholds for unit tests, for example, to avoid false test failures (type-I errors).
For example, we do not reject the hypothesis that [5060, 4940] comes from a uniform distribution:
>>> expected = { 0: 5000, 1: 5000 } >>> observed = { 0: 5060, 1: 4940 } >>> stochastic_equals(expected, observed, p=0.01) True
Here we also do not reject the hypothesis that a 6-sided die is unbiased:
>>> expected = { 1: 10, 2: 10, 3: 10, 4: 10, 5: 10, 6: 10} >>> observed = { 1: 5, 2: 8, 3: 9, 4: 8, 5: 10, 6: 20} >>> stochastic_equals(expected, observed, p=0.01) True
If we relax the p-value threshold, we can conclude that the die is in fact biased (but with some increased risk of type-I error):
But we would have if we used a 95% significance level instead of 99%: >>> stochastic_equals(expected, observed, p=0.05) False
leap_ec.util module
Defines miscellaneous utility functions.
TODO we have two almost identical counters that could be consolidated into a single class.
print_list : for pretty printing a list when pprint isn’t sufficient.
- leap_ec.util.birth_brander()
This pipeline operator will add or update a “birth” attribute for passing individuals.
If the individual already has a birth, just let it float by with the original value. If it doesn’t, assign the individual the current birth ID, and then increment the global, stored birth count.
We don’t increment a birth ID in the ctor because that overall birth count will bloat due to clone operations. Inserting this operator into the pipeline will ensure that each individual that passes through is properly “branded” with a unique birth ID. However, care must be made to ensure that the initial population is similarly branded.
Provides:
- brand_population() to brand an entire population all at once,
which is useful for branding initial populations.
brand() for explicitly branding a single individual
- Parameters
next_thing – preceding individual in the pipeline
- Returns
branded individual
- leap_ec.util.get_step(context={'leap': {'distrib': {'non_viable': 0}, 'generation': 100}}, use_generation=None, use_births=None)
Returns the current step of an algorithm using context. Will infer which to use if neither is specified with use_generation or use_births. If both are set to True, will raise an error.
- Parameters
context – the context from which the generations or births is taken from.
use_generation – an override to use generation.
use_births – an override to use births.
- leap_ec.util.inc_births(context={'leap': {'distrib': {'non_viable': 0}, 'generation': 100}}, start=0, callbacks=())
This tracks the current number of births
The context is used to report the current births, though that can also be obtained by inc_births.births()
This will optionally call all the given callback functions whenever the births are incremented. The registered callback functions should have a signature f(int), where the int is the new birth.
>>> from leap_ec.global_vars import context >>> my_inc_births = inc_births(context)
Each time we call the object, the birth count is incremented and returned:
>>> my_inc_births() 1
>>> my_inc_births() 2
>>> my_inc_births() 3
The count can be viewed without changing it like so:
>>> my_inc_births.births() 3
And decremented like so:
>>> my_inc_births.do_decrement() 2
- Parameters
context – will set [‘leap’][‘births’] to the incremented births
start – if we want to start counter at a higher value; e.g., take into consideration births of an initial population
callbacks – optional list of callback function to call when a birth numberis incremented
- Returns
function for incrementing births
- leap_ec.util.inc_generation(start_generation: int = 0, context={'leap': {'distrib': {'non_viable': 0}, 'generation': 100}}, callbacks=())
This tracks the current generation
The context is used to report the current generation, though that can also be given by inc_generation.generation().
This will optionally call all the given callback functions whenever the generation is incremented. The registered callback functions should have a signature f(int), where the int is the new generation.
>>> from leap_ec.global_vars import context >>> my_inc_generation = inc_generation(context)
- Parameters
context – will set [‘leap’][‘generation’] to the incremented generation
callbacks – optional list of callback function to call when a generation is incremented
- Returns
function for incrementing generations
- leap_ec.util.is_flat(obj)
- Returns
True if obj is a flat collection (as opposed to, say, a hierarchical list of lists).
>>> is_flat((0, 1)) True
>>> is_flat(1) False
>>> is_flat([(0, 1), (0, 1)]) False
- leap_ec.util.is_iterable(obj)
- Parameters
obj – that we want to determine is a generator
- Returns
True if obj can use next(obj)
- leap_ec.util.is_sequence(obj)
- Returns
True if obj is a test_sequence
Cribbed from https://stackoverflow.com/questions/2937114/python-check-if-an-object-is-a-sequence?lq=1
E.g., used to determine if gaussian mutation has a single specified standard deviation, or a vector of standard deviations.
>>> is_sequence(0.5) False
>>> is_sequence([0.1, 0.2, 0.3]) True
- leap_ec.util.print_list(l)
Return a string representation of a list.
This uses __str__() to resolve the elements of the list:
>>> from leap_ec.individual import Individual >>> import numpy as np >>> l = [Individual(np.array([0, 1, 2])), ... Individual(np.array([3, 4, 5]))] >>> print_list(l) [Individual<...> ..., Individual<...> ...]
As opposed to the standard printing mechanism, which calls __repr__() on the elements to produce
>>> print(l) [Individual<...>(...), Individual<...>(...)]
- Parameters
l –
- Returns
- leap_ec.util.wrap_curry(f)
Wraps and curries in conjunction, so the function signature remains.
Module contents
- leap_ec.leap_logger_name = 'leap_ec'
The environment variable we use to signal that our test harness is being run.